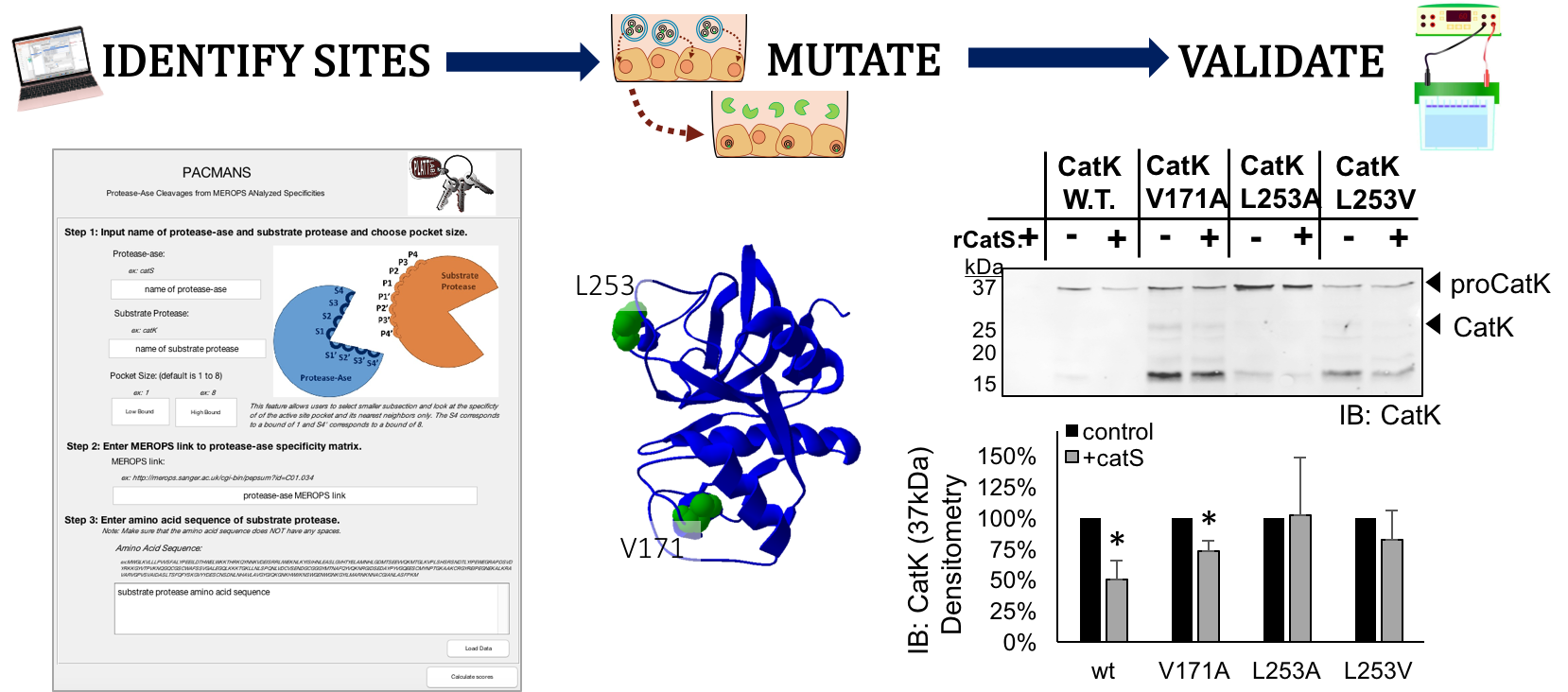

Multiple proteases in a system hydrolyze target substrates, but recent evidence indicates that some proteases will degrade other proteases as well. Cathepsin S hydrolysis of cathepsin K is one such example. These interactions may be uni- or bi-directional and change the expected kinetics. To explore potential protease-on-protease interactions in silico, a program was developed for users to input two proteases: 1) the protease-ase that hydrolyzes 2) the substrate, protease. This program identifies putative sites on the substrate protease highly susceptible to cleavage by the protease-ase, using a sliding window approach that scores amino acid sequences by their preference in the protease-ase active site, culled from MEROPS database. We call this PACMANS, Protease-Ase Cleavage from MEROPS Analyzed Specificities, and test and validate this algorithm with cathepsins S and K. PACMANS cumulative likelihood scoring identified L253 and V171 as sites on cathepsin K subject to cathepsin S hydrolysis. Mutations made at these locations were tested to block hydrolysis and validate PACMANS predictions. L253A and L253V cathepsin K mutants significantly reduced cathepsin S hydrolysis, validating PACMANS unbiased identification of these sites. Interfamilial protease interactions between cathepsin S and MMP-2 or MMP-9 were tested after predictions by PACMANS, confirming its utility for these systems as well. PACMANS is unique compared to other putative site cleavage programs by allowing users to define the proteases of interest and target, and can also be employed for non-protease substrate proteins, as well as short peptide sequences.